This page presents assignments used in a Parallel Computing course of the Computer Science degree, at the University of Valladolid, during the academic years from 2017/2018 to 2023/2024.
Table of contents
- Introduction
- Peachy Parallel Assignments
- Assignments description
- Download
- Acknowledgments
- Bibliography
Introduction
To parallelize applications, different programming models use different approaches. Understanding these variations is crucial for students to explore more advanced techniques and to tackle parallel programming in modern heterogeneous platforms effectively. These assignments serve as effective tools for teaching specific programming models and provide valuable insights into the portability of concepts and techniques across different models. Students can obtain a deeper understanding of the similarities and the conceptual shift between approaches, empowering students to critically analyze and select the most appropriate programming model and solutions for a given problem. The assignments can also be used with a single parallel programming model, exposing the main concepts needed to program effectively on any of them.
This collection includes six proposals of practical works, each covering a complete Parallel Computing or Programming course focusing on up to three programming technologies: OpenMP for shared memory, MPI for distributed memory, and CUDA/OpenCL for GPU accelerator programming. Each assignment in this series proposes one problem with a starting sequential code, to be parallelized and optimized by the students using the three supported technologies. The sequential program is intentionally designed to be straightforward and easily understandable for students, while it also provides specific parallelization and optimization opportunities. Each assignment covers the main competencies and abilities a student should develop in the parallel programming field such as loop parallelization and iteration scheduling; solving race conditions with critical regions, atomic operations, or reductions; data distribution and load balancing; collective vs. Point-to-Point communications; block size and geometry selection for GPU accelerators; optimization for locality and cache reuse; etc.
Each assignment includes a handout describing the problem, the sequential code, one version with the environment prepared to be parallelized with each programming technology, a makefile, and example input sets. The codes can be compiled with a debug mode activated that shows more information or even a graphical representation with ASCII art of the program evolution. The final output of the programs is some control and checksum information that can be used to check correctness easily. This information has been used during our courses to implement gamification strategies and programming contests with an online judge. Each assignment has been tested, corrected, and used during a Parallel Computing course, from 2017/2018 to 2022/2023, with a mean of 50 students on each course.
Peachy Parallel Assignments
Each assignment has been peer-reviewed and selected to be presented at the EduHPC workshop (Peachy Assignments), during Supercomputing conferences (SC2018, SC2019, SC2020, SC2021, SC2022, SC2023, SC2024, and SC2025) [1,2,3,4,5,6,7]. Peachy Assignments are a submission category at the EduPar and EduHPC conferences, and are peer-reviewed to ensure they meet the following criteria:
- Tested: All Peachy Parallel Assignments have been successfully used with real students.
- Adoptable: Peachy Parallel Assignments are easy to adopt. This includes not only the provided materials but also the content being covered. Ideally, the assignments cover widely taught concepts using common parallel languages and widely available hardware, have few prerequisites, and (with variations) are appropriate for different levels of students.
- Cool and inspirational: Peachy Assignments are fun and inspiring for students. They encourage students to spend time with the relevant concepts. Ideal Peachy Assignments are those that students want to demonstrate to their roommate.
Peachy Assignments are part of the Curriculum Initiative of NSF/IEEE-TPCC, involving the USA National Science Foundation, the Center for Parallel and Distributed Computing Curriculum Development and Educational Resources (CDER) in conjunction with the IEEE Technical Committee on Parallel Processing (TCPP).
You can find the proceedings papers describing our specific assignments through these links or those provided in the bibliography at the bottom of this page [1,2,3,4,5,6,7].
- Peachy parallel assignments at EduHPC’18
- Peachy parallel assignments at EduHPC’19
- Peachy parallel assignments at EduHPC’20
- Peachy parallel assignments at EduHPC’21
- Peachy parallel assignments at EduHPC’22
- Peachy parallel assignments at EduHPC’23
- Peachy parallel assignments at EduHPC’24
- Peachy parallel assignments at EduHPC’25 (to appear)
Assignments Description
2018 Energy Storms
Challege Rating: Easy
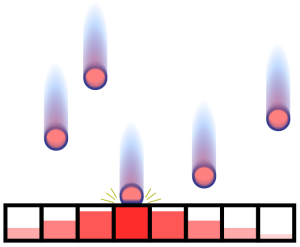
This assignment is based on a highly simplified simulation of the impact of high-energy particle storms on an exposed surface. It is inspired by simulations to test the robustness of space-vessel material. It uses 1-dimensional arrays to represent the discretized space. Each storm is represented as an unordered collection of pairs of numbers. Each pair indicates the impact position and the energy value of a particle. For each storm the program applies three stages: (1) Update the array cells depending on particles energy and distance to the impact point; (2) Compute a relaxation using a stencil operator; and (3) Compute a reduction to obtain the maximum energy value and its position in the array. The output of the program is the list of maximum values and positions after each storm.
2019 Fire extinghuising
Challege Rating: Intermmediate
This assignment is based on a simplified agent-based simulation where teams of firefighters aim to extinguish a set of fire focal points in a dynamically evolving scenario.
This assignment merges heat propagation (a classical iterative loop stencil computation) with an agent-based simulation, where agents are fire-extinguishing teams.
Two-dimensional arrays store the heat values on a discretized representation of the simulation arena. Fire focal points arise at established places and simulation times, and their heat is propagated to neighbor cells step by step. Teams of firefighters are represented by agents that make simple decisions to advance, one cell at a time, in the direction of the next nearest focal point to extinguish it. While teams advance, they also reduce the heat of neighbor cells in a given radius. There are three different team classes with different rules to move and different heat-reduction radii. The size of the simulation surface grid, the number and start frequency of focal points, and the number and class of the agents are the main dials for designing a new scenario with different load-balance properties.
In this animation, the numbers and symbols represent the heat accumulated at each point. Square brackets represent the position of fire-extinguishing teams. Normal brackets represent the fire focal points. We can see two teams extinguishing two fire focal points, the second one appearing when the two teams were already running towards the first. One of the teams changes the traveling direction and targets the second one. When the teams arrive at the focal points and extinguish them, the already propagated heat quickly decreases and disappears.
2020 Life evolution
Challege Rating: Intermmediate
This assignment is based on a simulation of life evolution, where protozoa learn to survive by getting food in a competitive environment. The idea is based on a simulation of the evolution of bugs feeding on bacteria in the muddy bottom of a stagnant pool of water by A. K. Dewdney [8]. The culture space is described by a 2-dimensional matrix where food values are stored. Some new food is randomly spread on each iteration, evenly or with a higher concentration in a specific zone. Some protozoa are randomly located at the beginning. The organisms are described by genes indicating the probability of advancing straight-forward or turning on their search for food. The food found by an organism is accumulated and spent each iteration to keep it alive and moving. When they reach maturity (a given number of simulation steps alive), if they have accumulated enough food, they may split into two new organisms. However, random mutations occur, and the genes of the descendants slightly change, modifying their moving behavior. During the simulation, we observe how the population grows and decreases, and how the organisms survive or starve. After enough iterations, the genes of the survivors present different features depending on the scenario parameters. The simulation results are determined by random seeds provided as arguments. Thus, they are reproducible. The input parameters can be chosen to generate specific situations with different growing and shrinking population sizes, different load distributions across the culture, different ratios of concurrency problems when protozoa collide in the same cell to get food, etc.
2021 Wind tunnel
Challege Rating: Advanced
This assignment is based on a simulation of the propagation of air pressure through a wind tunnel, in which obstacles modify the airflow, and moving particles are pushed. A lattice
represents a 2-dimensional cut of the tunnel space along the flow axis. In the upper row of the lattice there is a fan inlet. The air pressure is propagated in downward wavefronts at each simulation step using a directional stencil operator, leading to a pipeline computation.
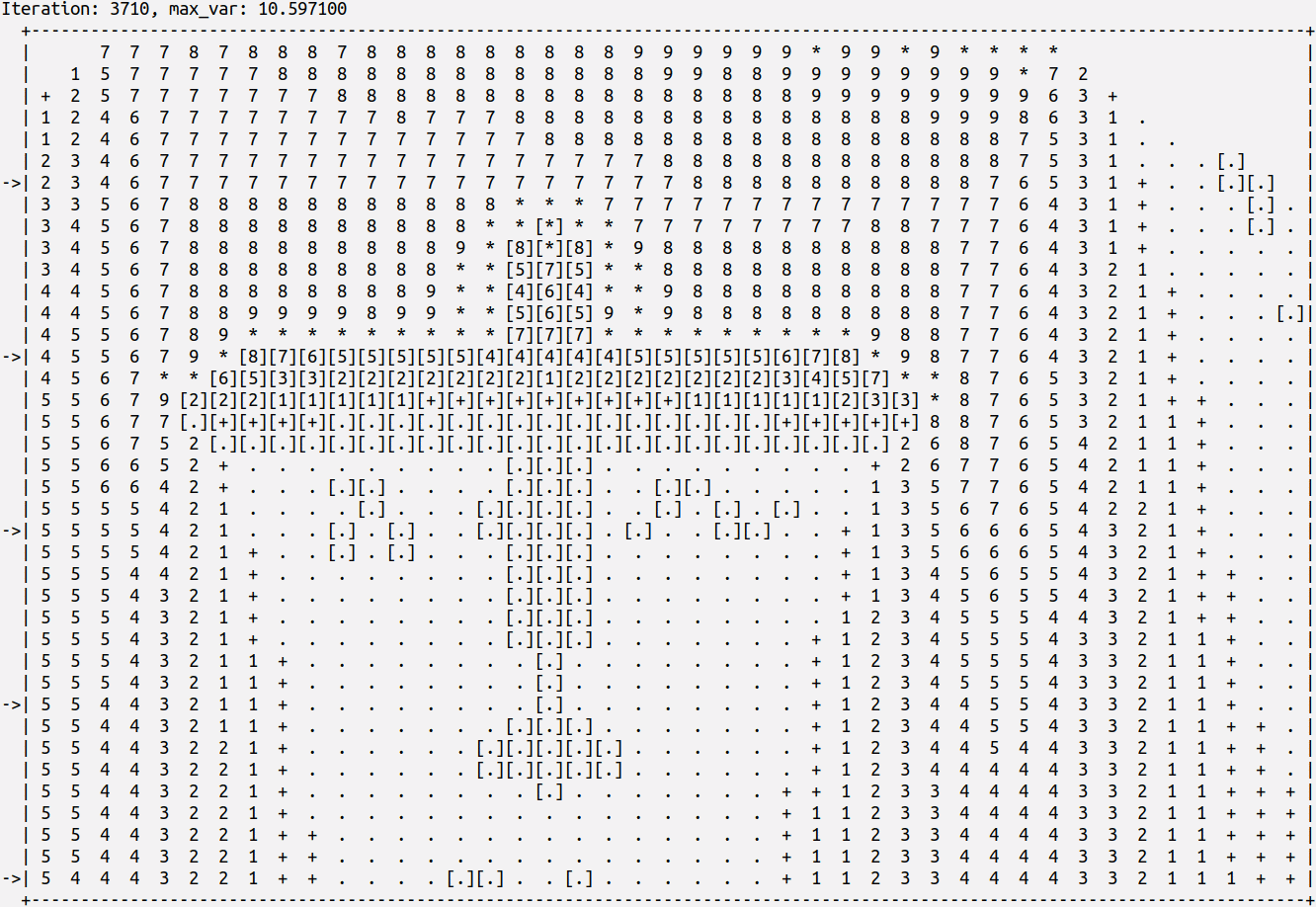
Fixed and flying particles can be added randomly at the start of the computation, or in chosen positions to form complex obstacles. For example, the shape of the airplane in the figure. Each particle has a given mass and airflow resistance. Different scenarios, with different obstacles and particle densities, can be easily created with the program parameters. The numbers and symbols in the figure represent the airflow pressure at each point. During the simulation, we can observe how the airflow is spread across the lattice and how the obstacles modify the air pressure. We can also observe how the flying particles are pushed by the airflow with simplified turbulent effects around the obstacles. The simulation results are determined by the initialization arguments that include random seeds. Thus, they are reproducible. The arguments can be chosen to generate specific situations with different load distributions, different ratios of concurrency problems when particles collide in a lattice position, etc.
2022 Hill Climbing with Monte Carlo
Challege Rating: Intermmediate
Multi-dimensional functions can model several physical systems, such as heat transfer, pressure flow, structural vibration, etc. Finding the values and locations of local and global maximums of such functions is important for the design and safety of many tools and physical structures in normal day-to-day life. Analytically locating the maximums of generic 2D functions can be hard. Thus, numerical approaches are used in many situations.
This assignment is based on a Monte Carlo probabilistic approach for a Hill Climbing algorithm, to locate the maximum values of a two-dimensional function in a parametric rectangular area. We provide an example function with multiple local maximums and other interesting features depending on the area selected. The selected area is discretized in a lattice with a parametric number of 2D control points. The hill-climbing approach is based on selecting a starting random point, and iteratively moving the selected point to one of the four neighbors: The one with the highest value of the function. When the selected-point value is higher or equal to the neighbor values, a local maximum has been reached and the search stops. The program uses a probabilistic approach to find the global maximum by starting a parametric number of hill-climbing searches. Each search can be executed in parallel, computing only the value of the function the first it is needed (memoization) and stopping the search when the path of another previous search is found. This introduces an interesting form of non-determinism, while the final results are predictable.
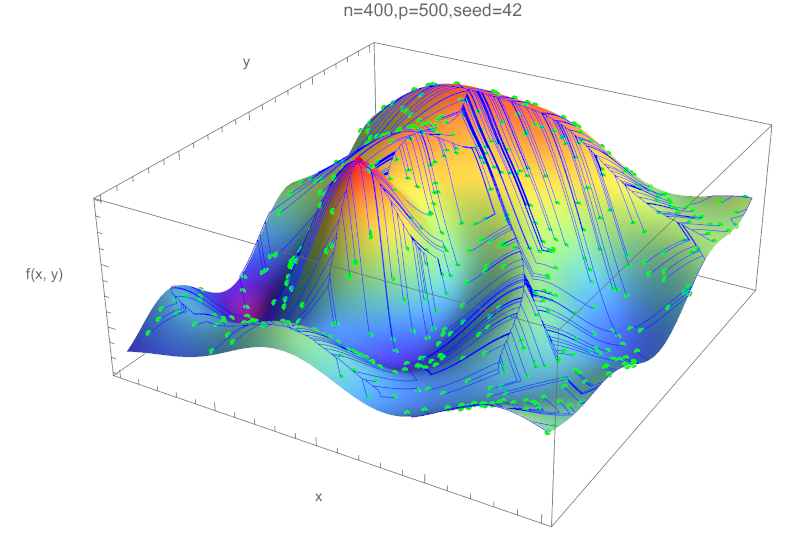
(Figure generated by the student Daniel García Solla)
2023 k-Means clustering
Challege Rating: Easy
The K-means clustering algorithm is a powerful and popular data mining algorithm used by the research community. It has applications in various fields, such as data segmentation, pattern analysis, image compression, or fault detection, among others. The code we provide to the students handles an initial cloud of points provided in a text file. An array structure is used to store the points, with as many rows as points and as many
columns as dimensions of the points. During the initialization stage, initial centroid positions are randomly fixed, one per cluster. The main clustering loop has two phases. First, each point is re-assigned to the cluster whose tentative centroid is at the shortest Euclidean distance. An index array is used to track the centroid assigned to each point. The code also tracks the number of cluster changes (points transitioning from one cluster to another). Second, a new centroid location is calculated for each cluster as the arithmetic mean of all the points assigned to that cluster.
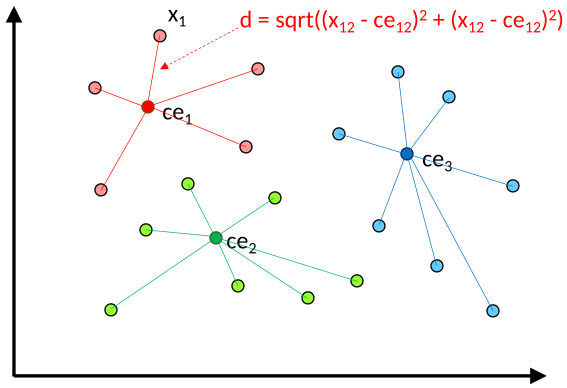
The figure shows an illustrative example of the cloud points assignment to centroids.
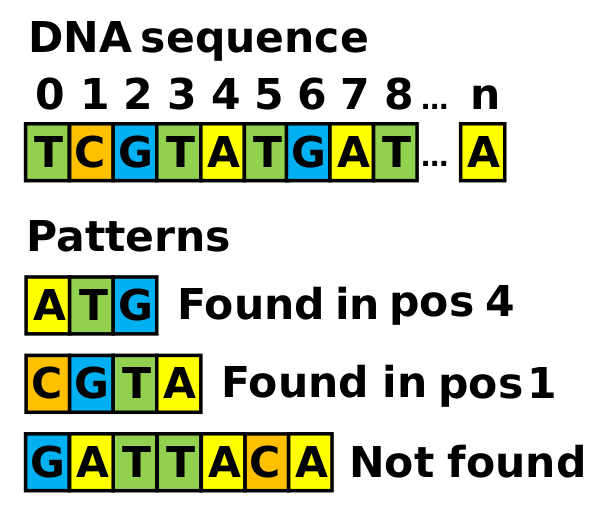
2024 DNA Sequence Alignment
Challege Rating: Easy / Advanced (MPI)
DNA sequences can be represented as strings of characters, where each character represents a nucleotide type. The exact DNA sequence alignment problem, or pattern searching problem, determines the exact coincidences of nucleotide strings in a long DNA sequence. The figure below illustrates an example of multiple pattern matching within a DNA sequence. Our assignment solves this problem for a random main DNA sequence and multiple nucleotide patterns that can also be random sequences and/or exact copies of parts of the main sequence randomly located. Several sophisticated algorithms have been proposed for this problem (see e.g. [9]). We choose a simple brute-force algorithm that checks each pattern starting at each possible position of the DNA sequence. It presents direct parallelization opportunities at two levels, patterns and starting positions. In our solution, if a pattern appears multiple times only the position of the first match is registered. In the sequential program, a pattern search stops when a match is found, skipping the search in the rest of the starting positions. The program determines which patterns are found in the sequence and their starting positions.
2025 Flood simulation
Challenge rating: Intermmediate
Floods can be modeled by simulating the accumulation of rainwater and its flow across a discretized surface. The height of the ground for each grid point can be represented with a value in a matrix. Our assignment provides four predetermined ground scenarios: a mountain terrain, a wide valley with hills, and two versions of the same valley with dams of different positions and heights. It includes a model to parametrize the generation of random collections of clouds that form fronts of a given size and depth. Individual clouds with fixed parameters can also be added. On each simulation step, the clouds advance across the scenario, dropping rain and increasing the water level of the grid points below the cloud. This is followed by a redistribution of water. For each matrix position, the amount of water that flows to the four neighboring positions is computed proportionally to the relative differences of their height and water level. The positions in the boundaries spill part of the water out of the matrix. In a second stage, each position is updated with the water coming from neighbors. This process is repeated for each iteration. This simulates the movement of water towards the lower areas of the terrain until the system reaches equilibrium or a maximum number of iterations has been computed. The video below illustrates an example of the simulation evolution.
Download
You can download the full assignments (including the handout, codes, makefile, and examples) at the following links:
- Energy Storms (EduHPC’18) (714 KB)
- Fire extinghuising (EduHPC’19) (147 KB)
- Life Evolution (EduHPC’20) (123 KB)
- Wind Tunnel (EduHPC’21 ) (148 KB)
- Hill Climbing with Monte Carlo (EduHPC’22 ) (122 KB)
- K-means (EduHPC’23) (14 MB)
- DNA sequence alignment (EduHPC’24) (118 KB)
- Flood simulation (EduHPC’25) (1.2 MB)

The material of these assignments is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Acknowledgments
This work has been developed in the context of GAMUVa (Grupo de Innovación Docente con Técnicas de Gamificación de la Universidad de Valladolid) partially funded by Vicerrectorado de Innovación Docente y Transformación Digital de la Universidad de Valladolid.
Bibliography
[1] Peachy Parallel Assignments (EduHPC 2018). E. Ayguadé, L. Alvarez, F. Banchelli, M. Burtscher, A. Gonzalez-Escribano, J. Gutierrez, D.A. Joiner, D. Kaeli, F. Previlon, E. Rodriguez-Gutiez, D.P. Bunde. Workshop on Education for High-Performance Computing (EduHPC 2018). Dallas (TX), USA, 2018. DOI: 10.1109/EduHPC.2018.00012. Proceedings 2018 IEEE/ACM Workshop for High-Performance Computing (EduHPC)
[2] Peachy Parallel Assignments (EduHPC 2019). M. Agung, M.A. Amrizal, S. Bogaerts, R. Egawa, D.A. Ellsworth, J. Fernandez-Fabeiro, A. Gonzalez-Escribano, S. Kundu, A. Lazar, A. Malony, H. Takizawa, D.P. Bunde. Workshop on Education for High-Performance Computing (EduHPC 2019). Denver (CO), USA, 2019. DOI: 10.1109/EduHPC49559.2019.00015. Proceedings 2019 IEEE/ACM Workshop for High-Performance Computing (EduHPC)
[3] Peachy Parallel Assignments (EduHPC 2020). H. Casanova, R.F. da Silva, A. Gonzalez-Escribano, W. Koch, Y. Torres, D.P. Bunde. Workshop on Education for High-Performance Computing (EduHPC 2020). Online event, 2020. DOI: 10.1109/EduHPC51895.2020.00012. Proceedings 2020 IEEE/ACM Workshop for High-Performance Computing (EduHPC)
[4] Peachy Parallel Assignments (EduHPC 2021). H. Casanova, R.F. da Silva, A. Gonzalez-Escribano, H. Li, Y. Torres, D.P. Bunde. Workshop on Education for High-Performance Computing (EduHPC 2021). St. Louis (MO), USA, 2021. DOI: 10.1109/EduHPC54835.2021.00012. Proceedings 2021 IEEE/ACM Workshop for High-Performance Computing (EduHPC)
[5] Peachy Parallel Assignments (EduHPC 2022). R. Carratalá-Sáez, A. Gonzalez-Escribano, A.S. Iliopoulos, C.E. Leiserson, I. Rosa, T.B. Schardl, Y. Torres, David P. Bunde. Workshop on Education for High-Performance Computing (EduHPC 2022). Dallas (TX), USA, 2022. DOI: 10.1109/EduHPC56719.2022.00012. Proceedings 2022 IEEE/ACM Workshop for High-Performance Computing (EduHPC)
[6] Peachy Parallel Assignments (EduHPC 2023). H.M. Bücker, J. Corrado, D. Fedorin, D. García-Álvarez, A. Gonzalez-Escribano, J. Li, M. Pantoja, E. Pautsch, M. Plesske, M. Ponce, S. Rizzi, E. Saule, J. Schoder, G.K. Thiruvathukal, R. van Zon, W. Weber, David P. Bunde. Workshop on Education for High-Performance Computing (EduHPC 2023). Denver (CO), USA, 2023. DOI: 10.1145/3624062.3625541. Proceedings 2023 IEEE/ACM Workshop for High-Performance Computing (EduHPC)
[7] DNA sequence alignment: An assignment for OpenMP, MPI, and CUDA/OpenCL. Arturo Gonzalez-Escribano, Diego García-Álvarez, Jesús Cámara. Workshop on Education for High-Performance Computing (EduHPC 2024). Atlanta (GA), USA, 2024. arXiv:2409.06075
[8] A. K. Dewdney. Simulated evolution: wherein bugs learn to hunt bacteria.
Scientific American, 260(5):138–141, May 1989.
[9] Smart exact string matching algorithm specifically for DNA sequencing. S. Ur Rehman and et.al.. 2nd International Conference on Cyber Resilience (ICCR). Dubai, United Arab Emirates. 2024.
